RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder | PNAS
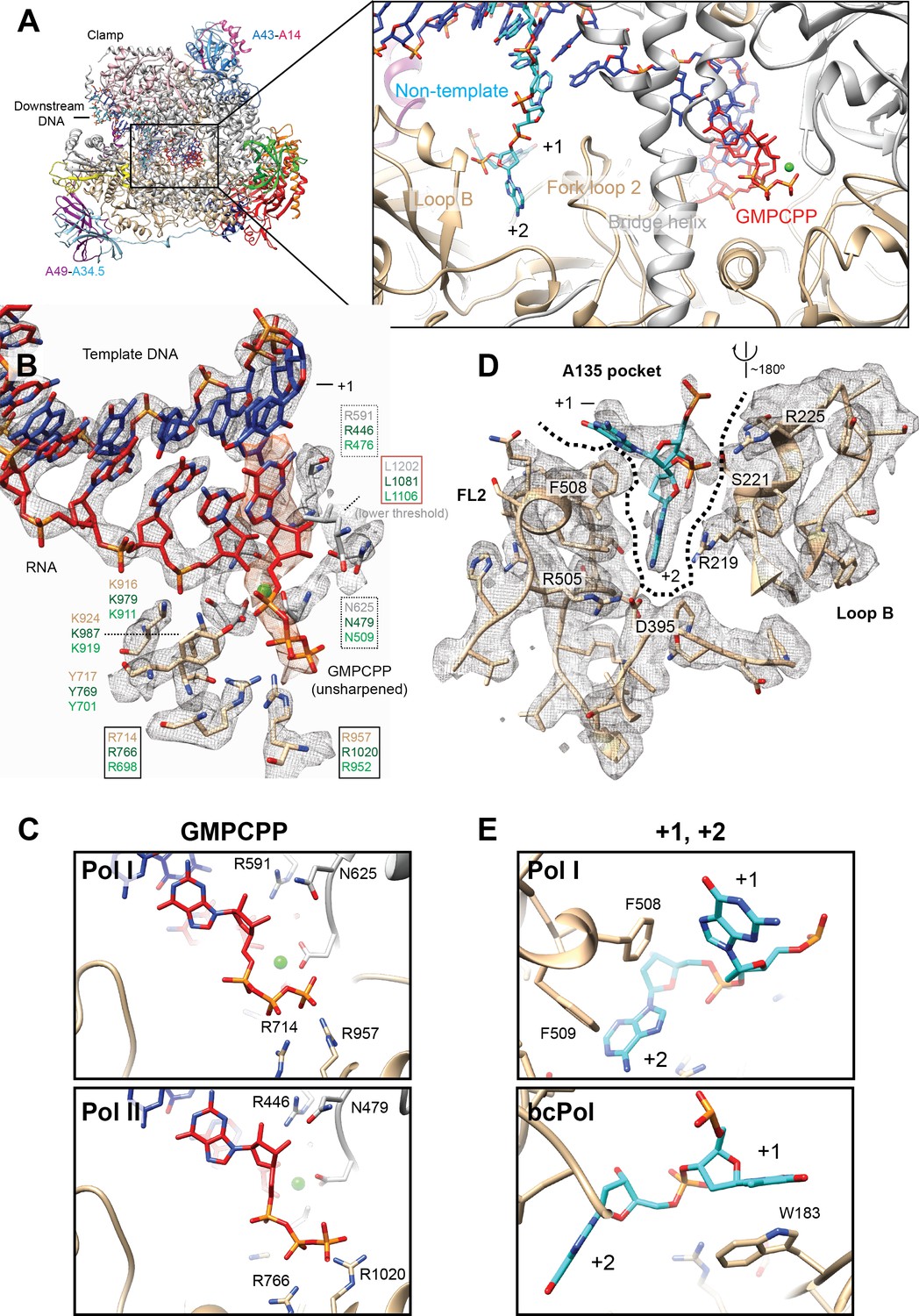
The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2 | eLife

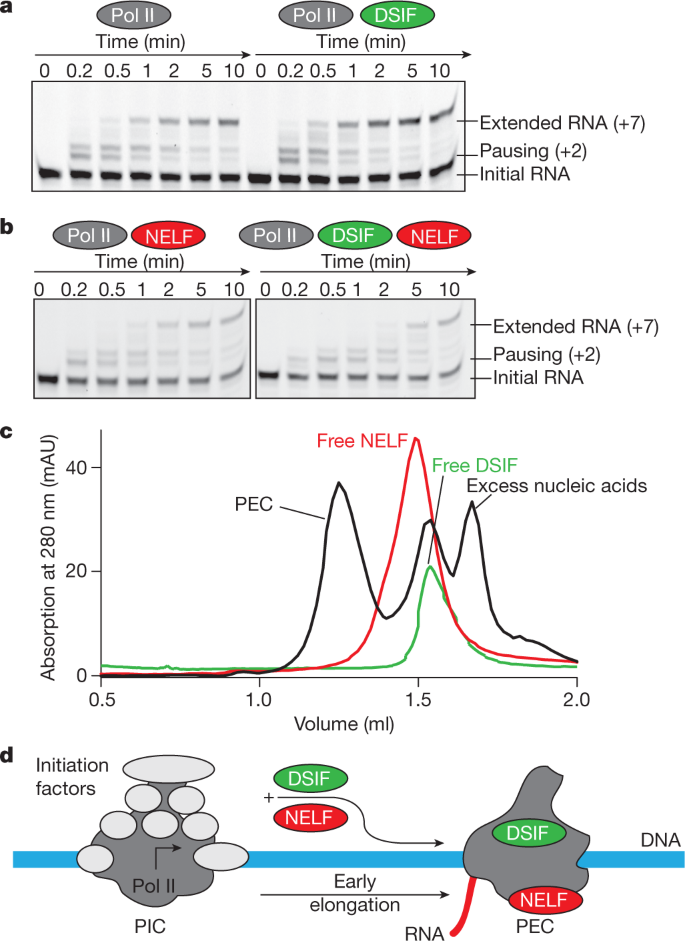
RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder | PNAS
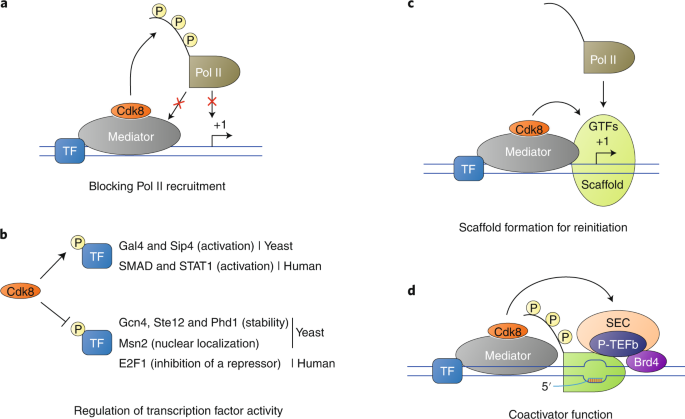
Dissecting the Pol II transcription cycle and derailing cancer with CDK inhibitors | Nature Chemical Biology

Roles of DNA polymerases V and II in SOS-induced error-prone and error-free repair in Escherichia coli | PNAS

Cell- and Polymerase-Selective Metabolic Labeling of Cellular RNA with 2′-Azidocytidine | Journal of the American Chemical Society

Phosphorylation-Regulated Binding of RNA Polymerase II to Fibrous Polymers of Low-Complexity Domains: Cell

RNA polymerase II transfer model. Depicted are steps involved in the... | Download Scientific Diagram

The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2 | eLife
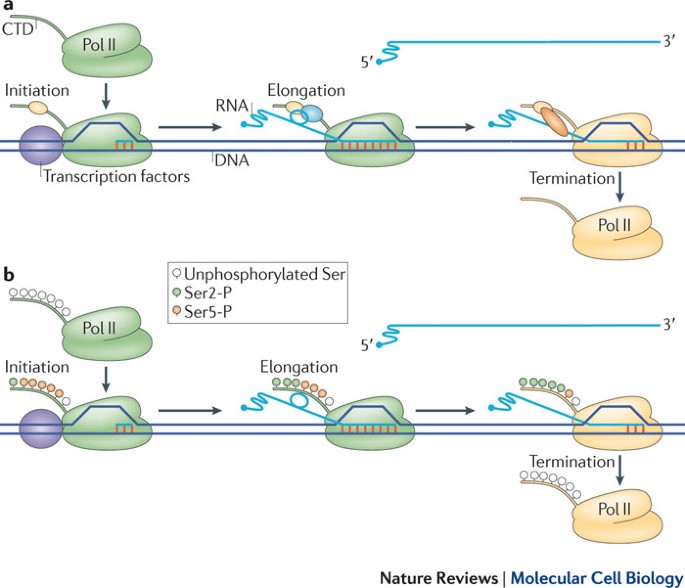
Unravelling the means to an end: RNA polymerase II transcription termination | Nature Reviews Molecular Cell Biology

RNA polymerase II clusters form in line with surface condensation on regulatory chromatin | Molecular Systems Biology